IX.4.2 Different genes in the same organism develop at different rates, even if the analysis only comprises selectively neutral traits
There are very substantial differences in the rate of the molecular clock between the individual genes of a single species.Sequences of pseudogenes change fastest, i.e. genes that have lost their function because of mutation in the regulation or coding region, and thus are not expressed in the cells or are expressed in the form of an inactive product.The untranscribed or untranslated regions of functional genes, including intron regions, exhibit high substitution rate.Exon regions develop most slowly (see Fig. IX.2).In genes coding, for example, ribosomal RNA, sections forming the double-helix secondary structure change most rapidly (Ben Ali et al. 1999), as only complementariness to the relevant base in the opposing parts of the chain and not the kind of pairing bases is generally important here from the standpoint of functioning of the molecule.
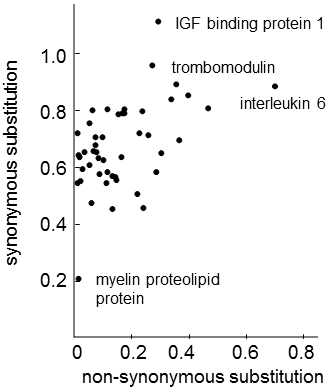
Fig. IX.7. Synonymous and nonsynonymous substitution in mammal genes. For 49 genes derived from rodents, primates and ungulates, the numbers of synonymous and nonsynonymous substitutions corresponding to one nucleotide were calculated. It is apparent from the graph that the substitution rate for synonymous substitutions is greater than the substitution rate for nonsynonymous substitutions and that there is smaller scatter in the individual genes in the number of synonymus substitutions than in the number of nonsynonymus substitutions. Simultaneously, there is a positive correlation between the two types of substitutions (p < 0.0001), where the number of nonsynonymous substitutions explains approximately 25% of the variability in the number of synonymous substitutions. Data according to Ohta (1995), modified according to Page and Holmes (2001).
It is apparent that the intensity and character of selection, to which the relevant gene or relevant section of the gene is exposed, is the decisive parameter affecting the substitution rate.Substitution rates differ substantially for the individual genes of a single organism and differences in these rates vary over a great many orders of magnitude.There are primarily very drastic differences in the substitution rates of nonsynonymous mutations.In contrast, the substitution rates for synonymous mutations vary in a much narrower range of values (Fig. IX.7).This is because the rate and probability of fixation of nonsynonymous mutations, i.e. mutations that lead to a change in the primary structure of the coded proteins and can thus be substantially manifested in changes in the fitness of their carriers, is determined by the intensity of the selection pressures active in the evolution of the relevant gene.These selection pressures understandably differ substantially for various genes (and various parts of these genes).However, there is a strong positive correlation between the rates of fixation of nonsynonymous and synonymous mutations.The source of this correlation is primarily the effect of genetic draft, a process that is also called thehitchhiking effect orgenetic hitchhiking.Because of the existence of this process, neutral mutations become fixed in the gene or are eliminated from the population together with selectively significant mutations (Braverman et al. 1995; Charlesworth & Guttman 1996).Thus, if a certain gene becomes the object of selection at a certain time, whether positive selection, i.e. towards evolutionary change, i.e. fixation of new mutations, or negative selection, i.e. towards evolutionary invariability, i.e. towards elimination of most new mutations, the rate of fixation of selectively significant and selectively neutral mutations is changed for it.The practical impact of this effect on the usefulness of the molecular clock in phylogenetics is that we must always attempt to analyze those genes that were not subject to changes in selection pressure in the past, even if we use only selectively neutral traits for the analysis.
It is necessary to emphasize that, although there is a certain dependence between the substitution rate for synonymous and nonsynonymous mutations in the context of the individual genes (Fig. IX.7), the rate of accumulation of neutral mutations in the context of the entire gene is certainly not directly related to the rate of anagenetic development (i.e. the development of phenotype properties, see XXIII.2) of the relevant species.Comparison of the sequence of genes for the α- and β-chains of haemoglobulin in the shark Heterodontus portusjacksoni, i.e. a chondrichthyes fish, whose morphology practically does not differ from that of some species of sharks that inhabited the Carboniferous seas 270 – 350 million years ago, indicated that the differences are comparable with the differences between the sequences of genes for the α- and β-chains of haemoglobulin in humans.This means that, in the phylogetic lines of sharks and humans, the same numbers of evolutionary substitutions in the α- and β-chains of haemoglobulin occurred from the moment of the formation of these genes through duplication of the originally single gene approximately 500 million years ago up to the present day (Kimura 1993).The results of study of the New Zealand tuatara (Sphenodon punctatus) yielded even more interesting results.Its appearance practically does not differ from that of its relatives living in the time of the Cretaceous; however, the molecular evolution of its mitochondrial DNA occurred much faster that in other species of vertebrates (Hay a spol. 2008).
