XI.1 All genes come from genes
Contemporary molecular biology has expanded the classical postulates of biology “All life comes from life” and “Every cell is formed from a cell” to include a third, similar postulate “All genes come from genes”.When sequences of various proteins were compared and especially when sequences of nucleic acids encoding various proteins were compared, it was found that there is lesser or greater similarity amongst the individual genes.This similarity can be most easily explained by the postulate that the individual genes were gradually formed from one another, i.e. that they are genealogically related (Ohno 1982).Where proteins perform similar functions, the similarity of their sequences is usually quite apparent; however, when they fulfill different functions, the similarity of some sections is usually completely or partly eliminated.We say that the sequences of the particular genes diverged, where the degree of their divergence is expressed as a percentage of different nucleotides or aminoacids in the same positions of the nucleotide or aminoacid chain.
Thus, genes form natural groups of mutually related genes called gene families (close relatedness) or superfamilies (distant relatedness), which can be ordered into genealogical trees using the mathematical methods of cladistics orphenetics, expressing the family tree of the relevant group of genes.The order of branching of these trees expresses the way in which evolution occurred of a certain family or superfamily, the time order in which the genes diverged from one another and the probable appearance of their common ancestor, the gene from which they developed.
About 750 protein families are known in the eukaryote, each of which has an average of 50 members.Proteins with more than 40% identity usually have the same function [10009].The best known example of large superfamilies of genes is probably the immunoglobulin superfamily.This superfamily includes a number of functionally quite unrelated genes; at random, we can mention the genes for immunoglobulins, T-cell receptors, class I and II MHC antigens, receptors for a constant part of immunoglobulins, Thy-antigen and a great many others (Fig. XI.1).
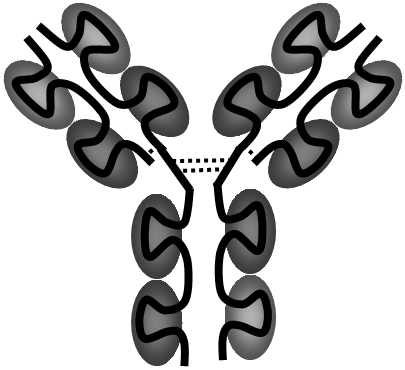
Fig. XI.1 Structure of a typical protein of the immunoglobulin superfamily. The scheme depicts the structure of the class IgG immunoglobulin molecule (secreted form). It consists of four chains of mutually covalently interconnected SS-bridges (dashed lines), where two light chains consist of two basic domains and two heavy chains consist of four domains. Variable domains, containing sections bonding the antigen, are depicted in a lighter colour, while the constant domains have a darker colour. The individual representatives of the immunoglobulin superfamily differ both in the number of chains forming their quaternary structure and also in the number of domains of which their chains are formed.
It can be expected that, when the sequences of other genes in modern organisms are elucidated, it will eventually be possible to order them into natural systems of mutually related families and superfamilies.The total number of these groups need not be very large.The genomes of eukaryotic cells contain approximately 10-40 thousand genes encoding various proteins (Lander et al. 2001).The current results of sequence analyses of a large number of genes simultaneously indicate that the number of structural elements, modules, from which these genes are composed, will apparently not be very great.Estimates of the number of unrelated exons, i.e. gene sections that are translated to their own protein chains, from which all contemporary genes are formed, vary around hundreds to thousands.This is not a very large number and it seems that exons will probably not be the basic module of the structure of genes, but rather composite structures, formed by combination of a smaller number of simpler modules [11620].Study of the structures of these gene modules and their evolutionary relationships will certainly constitute an extremely interesting and exciting chapter of molecular and evolutionary biology in the coming years.
