IX.4 Accumulation of neutral mutations in phylogenesis can be used as a molecular clock, allowing biologists to date phylogenetic events
Part of neutral mutations is usually gradually fixed in the gene pool of a particular biological species through genetic drift.As the frequency of fixation of neutral mutations in time depends only on the mutation rate, of which it is assumed that it is roughly constant during phylogenesis for most organisms, it is possible for biologists to determine the time that has expired from the moment of divergence of two sister groups (taxa) from the common exclusive ancestor on the basis of the number of substitutions that occurred independently in the two lines from the moment of divergence.If we sequence a certain DNA section for two related species and determine the number of neutral mutations in which they differ, on the basis of a mathematical model that takes into account and eliminates the effect of possible repeated mutations in the same position, we can estimate how many fixation events occurred in the two species from the moment when they branched off from the common ancestor.If, in addition, we know the characteristic substitution rate for the given taxon and the given gene, i.e. the average number of mutations fixed for the given species per time interval, then we can calculate the time that has elapsed since the branching off of the relevant phylogenetic lines.Thus, fixation of neutral
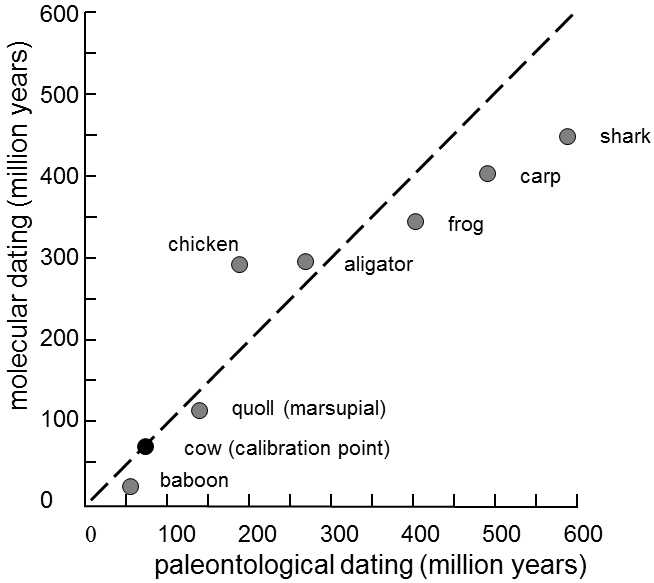
Fig. IX.5. Calibration of the molecular clock. The age of the branching-off of the line leading to humans from the line leading to the other vertebrates, determined on the basis of paleontological data, is plotted on the abscissa, while the same age determined on the basis of accumulation of substitutions in the gene for α-globulin is plotted on the ordinate. Calibration of the α-globulin molecular clock was performed on the basis of branching off of the lines leading to humans and to cattle, assuming that this event occurred 80 million years ago. It is apparent from the graph that, for most of the studied vertebrates, reasonable agreement exists between the dated moments of branching off of the individual species on the basis of paleontological data and on the basis of the molecular clock. Modified according to Page and Holmes (2001).
mutations can act as a molecular clock, permitting more or less exact dating of the individual events in phylogenesis or, to be more exact, the individual splitting events that occurred during the cladogenesis of the studied taxon.
It is obvious that this substitution rate is frequently not known.However, in this case, we can calibrate the molecular clock on the basis of the number of evolutionary changes in which the two studied species differ from a third species for which we know the moment of divergence from the paleontological record (Fig. IX.6).If, for example, we know that the taxon including species A and B branched off from the taxon including species C at time T1 ago and, since that time, species A has collected KACmutations in the studied gene and species B KBCmutations, where, since the time of splitting off of species A and B, i.e. over time T2, species A and B collected KABmutations, we can calculate the time expired since divergence of species A and B according to the equation
T2 = (2KABT1)/(KAC + KBC).
If, on the other hand, we know the time that has expired since branching off of species A and B and we are interested in the time that has expired since the splitting off of these species from species C, we can use the equation
T1= (KAC + KBC) T2/2KAB.
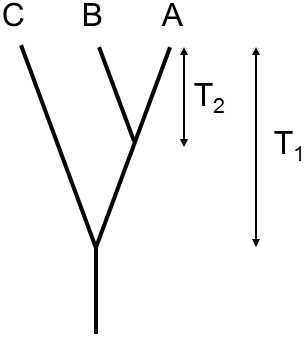
Fig. IX.6. Calculation of the moment of branching off of the line using the molecular clock. On the basis of the number of substitutions in the individual branches of the phylogenetic tree and knowledge of the age of one of the points on the branching off of three taxons, it is possible to simply estimate the age of the second point on the branching. If molecular evolution occurred at a different rate in the various branches, however, our estimates can be erroneous.
